DEBtox information
Making sense of ecotoxicity test results

Main menu
Submenu
| |
||
 DEBtox information Making sense of ecotoxicity test results |

|
|
|---|---|---|
Main menu |
Submenu |
|
DEBtox information |
DEBtox information |
|
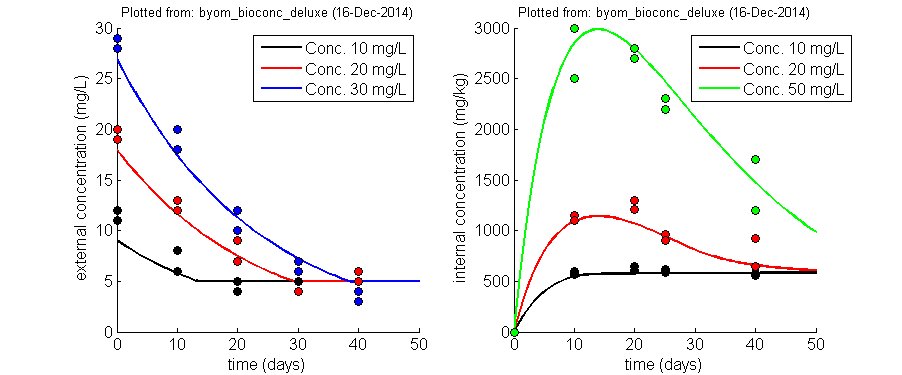
BYOM flexible model platformBring/Build Your Own Model (BYOM) is a flexible set of Matlab scripts and functions to help you build, simulate and fit your own models. Not just DEB models or TKTD models; any model that you want (well, as long as it can be expressed in terms of ordinary differential equations, or as explicit function). Using these files requires a working knowledge of Matlab, as there is no nice GUI. If you're new to Matlab, check out the free tutorials that the MathWorks offers, or some of the on-line course of Udemy (not free). I originally created this platform for teaching purposes: I wanted students to build and fit their own models, but I wanted them to focus on the modelling and not on the programming (BYOM takes most of the difficulties away from the user). However, I find myself using it almost exclusively now, as it is a perfect platform (for me) for both standard and especially non-standard (one of a kind) modelling. BYOM is also used in the TKTD summercourse in Denmark. Please note that user-friendliness is limited, which is generally the consequence of high flexibility. You need to be able to work with Matlab at a fairly basic level, and get used to the BYOM format, to be able to work with it effectively. The TKTD summercourse is a good place to get started with it. BYOM and the additional packages are totally free (but Matlab is, of course, not). If you like BYOM, let me know! You can support maintenance and further development of BYOM by buying one of my e-books. Below, an example fit with BYOM (simultaneous fit on two state variables, with replicated data for different concentrations and different observation times) |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 FEATURES:
---> Download BYOM_v69.zip for Matlab (version 6.9, 9 April 2024) --> version_log ---> I have been working on an on-line manual for BYOM. You can look at the in-progress version that already covers the BYOM basics (updated 15 Nov. 2021). Whenever I have time I will update it and add sections to it, so in the end it will hopefully cover all of the nooks and crannies of BYOM. You may also download the PDF manual BYOM 3.x (3 April 2015). This manual will eventually be completely replaced by the on-line manual. ---> Walk through the BYOM code to see what it does! (this is for version 6.0 BETA 6) LICENSE INFORMATION. The BYOM implementation and the associated packages are distributed under an MIT-style permissive license. The license can be downloaded here, and is included in the BYOM zip file as well. From v. 5.0, the parameter-space explorer from openGUTS was modified to be used in BYOM as well. These functions are in a sub-function of the engine folder named <parspace> and are distributed under the terms of the GNU General Public License (just like the openGUTS files from which they were derived). OLD VERSIONS. The
BYOM versions as well as the packages are archived, by me,
off-line. That is done since the official release of
version 1.0 in 2012. Therefore, if you need an older
version of the software, for example to re-run an old
analysis, please contact
me and I can send you the files. Disclaimer and warnings!
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name |
Description |
Version
and date |
Download |
| GUTS |
This package contains both simple
and more elaborate GUTS
implementations. Easy to adapt, and can accommodate
time-varying concentrations. Includes a script to
produce synthetic data sets. Take a look at the walkthrough
(for v. 2.3). |
version
3.3 21 Sept. 2023 |
GUTS_v33.zip |
| DEBkiss |
A general version of the DEBkiss model, so more useful as starting point for your own analyses. Take a look at the walkthrough (version 2.3a). | version
2.3a 30 Nov. 2021 |
DEBkiss_v23a.zip |
| DEBtox classic |
Classic simplified DEBtox model.
This is the version as published by Jager
& Zimmer (2012). Same model as in DEBtoxM
flavour "Basic DEBtox", but with more flexibility.
Includes case studies for Capitella and Daphnia
that also demonstrate the calculation of the
intrinsic rate of population increase. Take a look
at the walkthrough
(version 2.3a). Note: this package is NOT up-to-date
with the newest insights, as included in the DEBtox e-book (version 2.0
and newer) and the revisited DEBtox2019 (Jager,
2020), which place 'damage' in a central
position. The DEBtox2019 package below IS fully
updated, and is my advice for a 'standard'
simplified DEB-TKTD model. |
version 2.4 22 Sept. 2023 |
DEBtox_classic_v24 |
| SIMbyom |
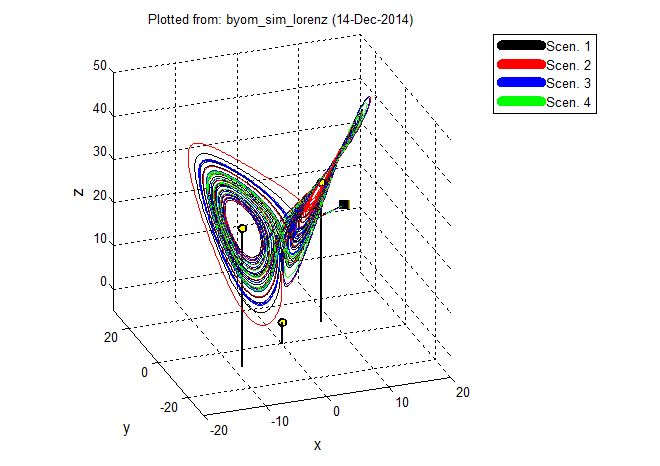
A simulation tool for BYOM models. Can plot state vs. time, but also state vs. state (2d and 3d), or derivative vs. state. Several example systems are provided: two chaotic systems, log-logistic growth, and the Rosenzweig-MacArthur population model. Take a look at the walkthrough (version 2.1). | version
2.1 17 Nov. 2021 |
SIMbyom_v21.zip |
| DEBtox2019 |
Updated simplified DEBtox model,
based on DEBkiss.
A paper presenting and explaining this version is
published (Jager,
2020). This version is largely equivalent to
the one described in the DEBkiss e-book
version 2 (and completely equivalent to upcoming
version 3), and includes the simplified model
versions as treated in Chapter 6. This version
brings DEBtox in line with the developments in GUTS.
The package includes case studies for Folsomia,
Capitella and Daphnia. Take a look
at the walkthrough
(for version 4.5a). With version 4.6, the models
using primary parameters are moved to the new
package DEBkiss_tox. More
information on this package. See also the
associated DeEP tool to
make predictions from a DEBtox2019 calibration to
long-term exposure profiles. Version 4.7 of the
package (ERA-special folder) shows how BYOM
calibration results can be converted to DeEP input
files. |
version
4.7 13 Dec. 2022 |
DEBtox2019_v47.zip |
| doseresp |
Use BYOM to fit a simple log-logistic
dose-response curve to quantal (e.g., survival) or
graded (e.g., reproduction) data. For quantal data,
the binomial distribution is used. Now you can use
BYOM tools like profiling and Bayes to your classic
dose-response analyses (but see warning over here). Take a
look at the walkthrough
(version 1.3). |
version
1.3 17 Nov. 2021 |
doseresp_v13.zip |
| GUTS_mix |
Extension of the reduced GUTS models
SD and IT for dealing with binary mixtures. The
model extension is presented and discussed in Bart
et al (2021). More
information on this package. This package
requires BYOM v.6.0 beta 8 or newer! |
6 Dec. 2021
|
GUTS_mix_v10.zip |
| stdDEB-TKTD |
Implementation of standard DEB with
the TKTD module from DEBtox2019. This package
requires use of AmP parameters for the species. In
general, I would recommend using DEBtox2019, though,
because of its simplicity. This package requires
BYOM v.6.0 beta 8 or newer! This package needs a bit
more testing, especially for pulsed exposure, and it
is a rather complex piece of code. The paper
presenting this model version is Jager
et al (2023). |
version
1.1 8 March 2023 |
stdDEB_tktd_v11.zip |
| DEBkiss_tox |
Updated simplified DEBtox model, based on DEBkiss.This package was split out of the DEBtox2019 package, and contains the models in primary parameters (with either primary parameters or compound parameters as input). In general, I advise to use the DEBtox2019 package. This formulation is for further experimentation with additional pMoAs or additional endpoints. More information on this package. | version
1.0 16 Sept. 2022 |
DEBkiss_tox_v10.zip |
| DEBtox_mix |
An experimental package for
DEBtox-based binary-mixture analysis. The model
follows DEBtox2019 (Jager
2020). This package requires BYOM v.6.8 or
newer! More
information on this package. |
version
1.0 18 Sept. 2023 |
DEBtox_mix_v10.zip |
| GUTS_immob |
An experimental package for GUTS
immobility, expanding GUTS to distinguish between
(reversible) immobility and (irreversible) death. A
publication explaining the model is upcoming (will
be linked when published). This package is very
difficult to use as there are many options to
configure the model, and because many parameters
need to be fitted. Also, calculations are very time
consuming. So this package is recommended for
experts only. This package requires BYOM v.6.9 or
newer. |
version
1.0 9 April 2025 |
GUTS_immob_v10.zip |
| Name |
Description |
Version
and date |
Download |
| DEBkiss-paper | This package contains the files to
reproduce the figures for the pond snail in the
DEBkiss paper (Jager
et al., 2013). It is a basic implementation of
DEBkiss, for continuous reproduction only, with
optional maturity maintenance. Several things are
specific for this example, so use the DEBkiss
package above for your own analyses. Take a look at
the walkthrough
(older version). |
version 2.1 29 Nov. 2021 |
DEBkiss-paper_v21.zip |
| support-marecotox |
Support package to reproduce the case
studies in the chapter on modelling in the book "Marine Ecotoxicology". |
version
2.2 1 Dec. 2021 |
support-marecotox_v22.zip |
| Acute Calanus package |
This package contains the files
needed to reproduce the results of the paper
"Dynamic links between lipid storage, toxicokinetics
and mortality in a marine copepod exposed to
dimethylnaphthalene." Environ Sci Technol http://dx.doi.org/10.1021/acs.est.7b02212
(standard GUTS analyses, TK analysis with a one- and
two-comp. model, and combined GUTS and two-comp. TK
analysis). |
version 1.2 2 Dec. 2021 |
acute_calanus _package_v12.zip |
| GUTS delayed mortality |
This package contains the files
needed to reproduce the analyses in an upcoming
paper on delayed mortality. The packages contains a
text file with notes on how to reproduce the
analyses. |
5 Dec. 2023
|
GUTS_delayed _mort_v10.zip |